Computational Methods for Cancer and Drug Discovery
Molecular subtypes of ovarian cancer
In this project, I sought to leverage a compendium of whole-transcriptome gene expression datasets to better understand molecular subtypes of high-grade serous ovarian carcinoma. I developed a random forest-based model, consensusOV, that outputs subtype predictions and real-valued posterior probabilities, and demonstrated an emerging consensus among independently published proposed subtypes. This is available in the R package consensusOV.
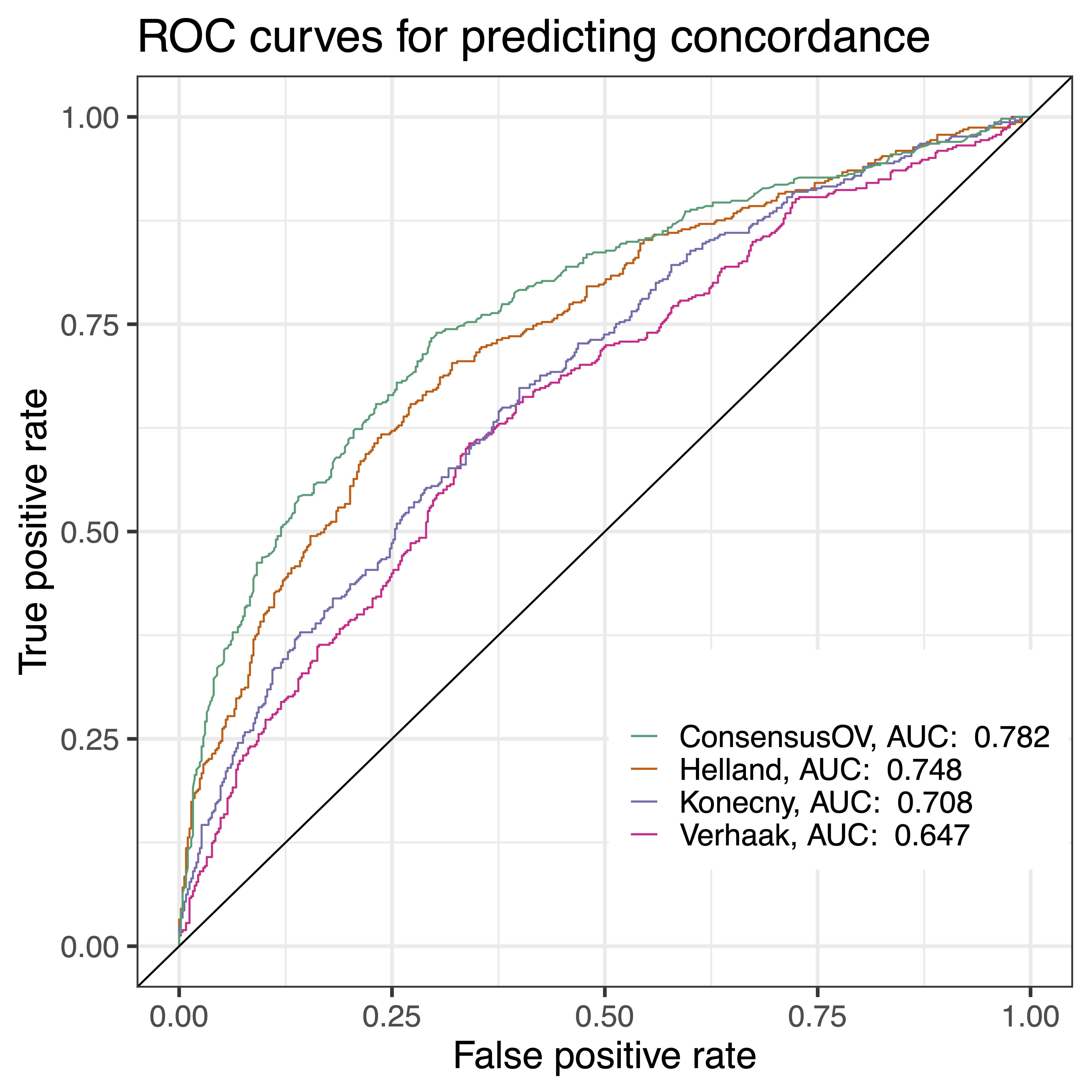
Chen GM*, Kannan L*, Geistlinger L, Kofia V, Safikhani Z, Gendoo DM, Parmigiani G, Birrer M, Haibe-Kains B#, Waldron L#. Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma.
Clinical Cancer Research (2018).
Computational methods for drug discovery
A significant proportion of small-molecule drugs derive from natural products produced by microorganisms such as bacteria and fungi. These organisms produce bioactive small molecules through a fascinating system of biosynthetic machinery, in which their genomes encode enzymatic ‘assembly lines’ that synthesize chemical structures in a stepwise fashion. One of the key challenges in this field is: how can we connect the large quantities of genomic data with chemical structures and biochemical knowledge to improve the process of discovering new drugs? Toward this challenge, I designed two computational tools: GRAPE, a tool for predicting the biosynthetic assembly lines involved in a given small-molecule chemical structure; and GARLIC, a dynamic programming algorithm for matching small molecule chemical structures to the underlying biosynthetic genomic components. Along with other talented scientists in the Magarvey Lab, these tools have been used to link natural products to ‘orphan’ biosynthetic gene clusters, and identify an antibiotic family with a new mechanism of action.
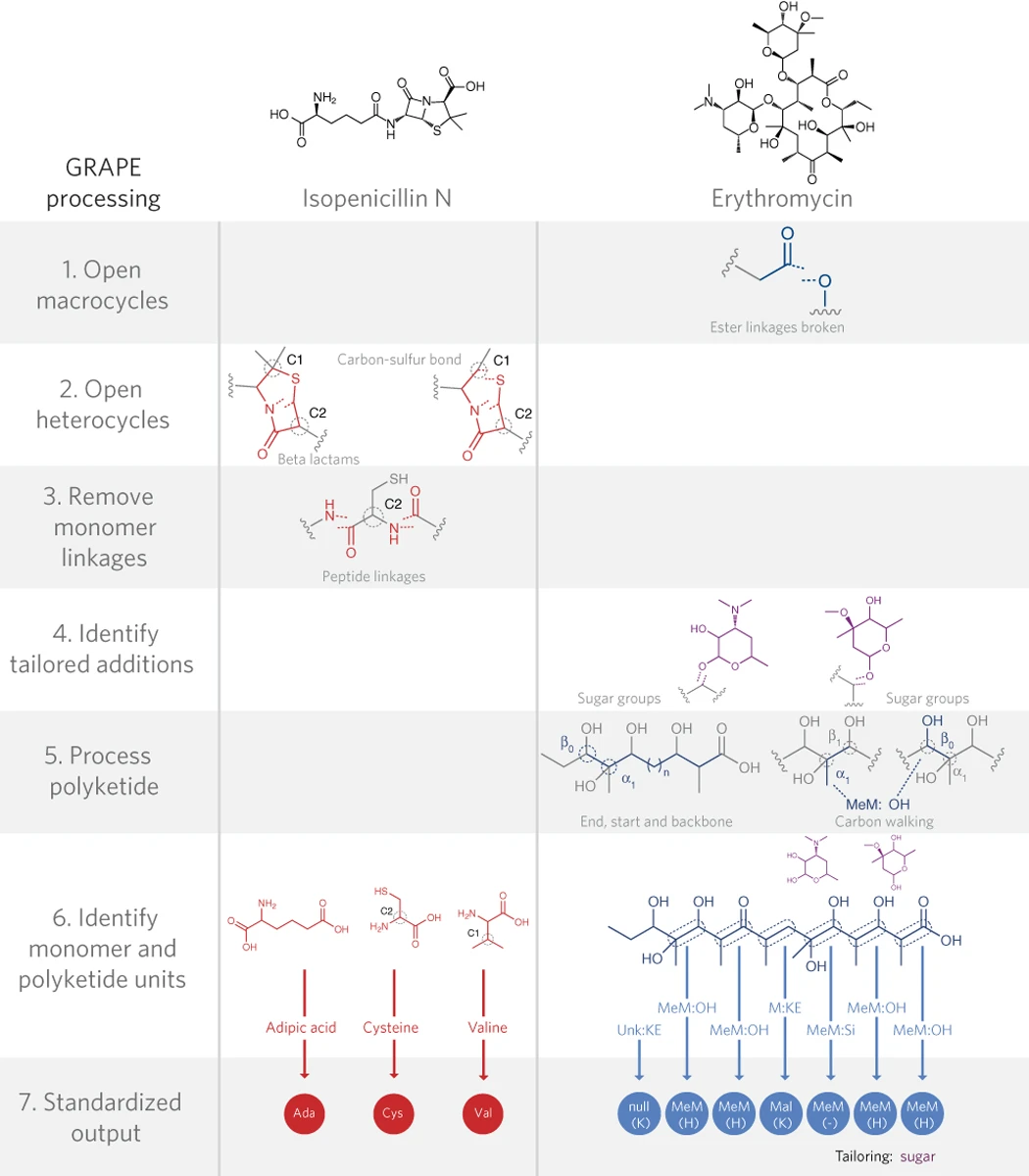
Dejong CA*, Chen GM*, Li H*, Johnston CW, Edwards MR, Rees PN, Skinnider MA, Webster ALH, Magarvey NA. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching.
Nature Chemical Biology (2016).
Johnston CW, Skinnider MA, Dejong C, Rees P, Chen GM, Walker C, French S, Brown ED, Berdy J, Liu D, Magarvey NA. Collecting and clustering natural antibiotics guides identification of evolved targets.
Nature Chemical Biology (2016).